Researchers in the Phan Group at the Latvian Institute of Organic Synthesis, published in Communications Chemistry, have characterized a serine protease that cleaves precursor peptides with unprecedented uniformity. The enzyme WprP2 from Streptomyces venezuelae recognizes two distinct cleavage sites regardless of intervening sequence length or amino acid composition, expanding the toolkit for processing ribosomally synthesized and post-translationally modified peptides into bioactive natural products.
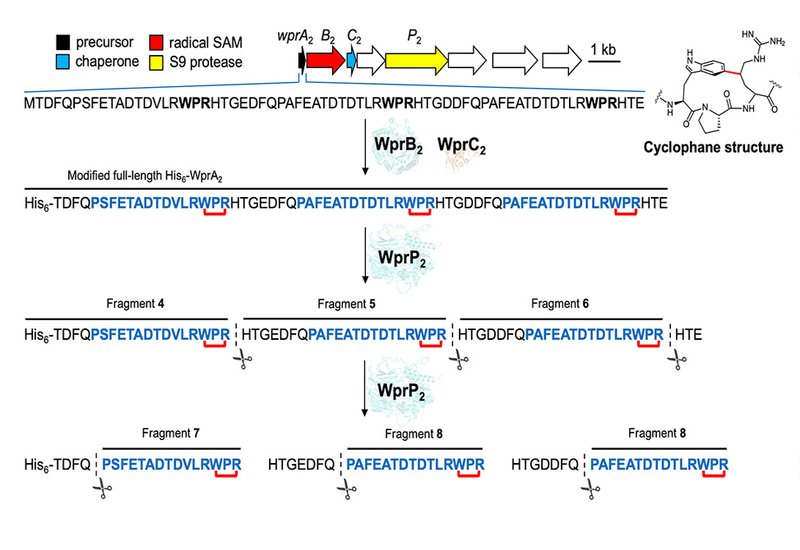
Ribosomally synthesized and post-translationally modified peptides represent a rapidly growing class of natural products defined by their diverse chemical modifications. Their biosynthesis requires proteases to remove leader peptides from precursor sequences, releasing mature bioactive molecules. The S9 protease family has gained attention for its role in processing these peptides, yet only three members have been characterized in this context: FlaP, OphP, and MpgP. These enzymes participate in lanthipeptide, omphalotin, and clavusporin biosynthesis, respectively. The team identified WprP2 from Streptomyces venezuelae NPDC049867, encoded adjacent to the radical SAM enzyme WprB2 involved in cyclophane natural product biosynthesis. Cyclophane-containing peptides such as darobactin A have attracted significant interest for their activity against Gram-negative bacteria. Understanding how WprP2 recognizes and cleaves its substrates could enable new approaches for generating bioactive peptides with cross-linked architectures.

The Phan Group at the Latvian Institute of Organic Synthesis
From left to right: Chin-Soon Phan, Anitra Zīle, Gaja Swarna Kumari, Abujunaid Habib Khan, Vic Kiselov, and Jabal Rahmat Haedar, first author of this paper.
The researchers expressed WprP2 in Escherichia coli and tested its activity against precursor peptides containing three WPR sequence motifs. Liquid chromatography-mass spectrometry revealed that the enzyme cleaves after all three WPR motifs uniformly, generating distinct peptide fragments. Further analysis uncovered a second cleavage event: the protease also cuts before proline residues at the twelfth position preceding each WPR motif when glutamine precedes the proline. Mutational studies pinpointed the recognition sequences as WPR-Xxx for the first cleavage and Gln-Pro for the second. The team confirmed the catalytic mechanism through alanine substitutions at the catalytic triad residues Ser507, Asp590, and His621, which abolished activity entirely. AlphaFold3 structural predictions positioned both cleavage sites near this catalytic triad, supporting the proposed sequential processing model. Time-course experiments demonstrated that longer peptide fragments appear before shorter ones, consistent with stepwise cleavage at multiple sites.
WprP2 differs from characterized S9 proteases in three key ways: it does not cluster phylogenetically with FlaP or OphP, it cleaves at the N-terminus of proline rather than after it, and it processes multiple sites uniformly across the precursor peptide. The enzyme also cleaves unmodified substrates with equal efficiency, indicating that cyclophane installation by the partner radical SAM enzyme is not required for substrate recognition. This promiscuity in accepting substrates with varied amino acid compositions and lengths between cleavage sites suggests broad applicability for peptide engineering. The researchers tested WprP2 against a related precursor peptide with different intervening sequences and observed consistent cleavage at WPR motifs. When they introduced a glutamine before proline through mutagenesis, the second cleavage activity appeared as predicted. Commercial trypsin failed to cleave the same WPR-His site that WprP2 processes efficiently, highlighting the enzyme's specialized utility for peptide bond cleavage in contexts where standard proteases fall short. This work adds to the growing repertoire of RiPP proteases and demonstrates the potential of WprP2 for generating bioactive peptides through its unique uniform cleavage activity.